本文主要是介绍【Segment Anything Model】十三:Meta的最新工作EfficientSAM,微调到自己的数据集,代码。,希望对大家解决编程问题提供一定的参考价值,需要的开发者们随着小编来一起学习吧!
🍉 博主微信
cvxiayixiao还有其他专栏点击头像查询
🍓 【Segment Anything Model】计算机视觉检测分割任务专栏。
🍑 【公开数据集预处理】特别是医疗公开数据集的接受和预处理,提供代码讲解。
🍈 【opencv+图像处理】opencv代码库讲解,结合图像处理知识,不仅仅是调库。
文章目录
- 1️⃣预备知识
- EfficientSAM要解决的问题
- EfficientSAM解决此问题的创新点
- 知识蒸馏和利用掩码图像去做预训练
- 架构解释:
- 上半部分作用?
- 上半部分采用的方法
- 下半部分作用?
- EfficientSAM的结果
- 2️⃣EfficientSAM用于自己的数据集代码
- 处理数据集
- 将Efficient-SAM代码和权重拷贝到服务器或者本地
- 去官网git下载权重
- 愉快训练
- 单卡
- 多卡
1️⃣预备知识

EfficientSAM要解决的问题
sam本身架构庞大,训练和推理都很慢。
EfficientSAM解决此问题的创新点
知识蒸馏和利用掩码图像去做预训练
具体来说:
- 利用掩膜图像预训练(SAMI)来学习从SAM图像编码器中重构特征,以有效进行视觉表示学习。这是提高EfficientSAMs效率和性能的核心策略。
- 蒸馏到轻量级图像编码器和掩码解码器: 采用SAMI预训练的轻量级图像编码器和掩码解码器构建EfficientSAMs,进一步降低模型复杂度,同时保持良好的性能。
- 在SA-1B数据集上的微调: 经过简化的架构在SA-1B上微调。还可以继续做下游任务,包括:
图像分类、对象检测、实例分割和语义对象检测。
架构解释:
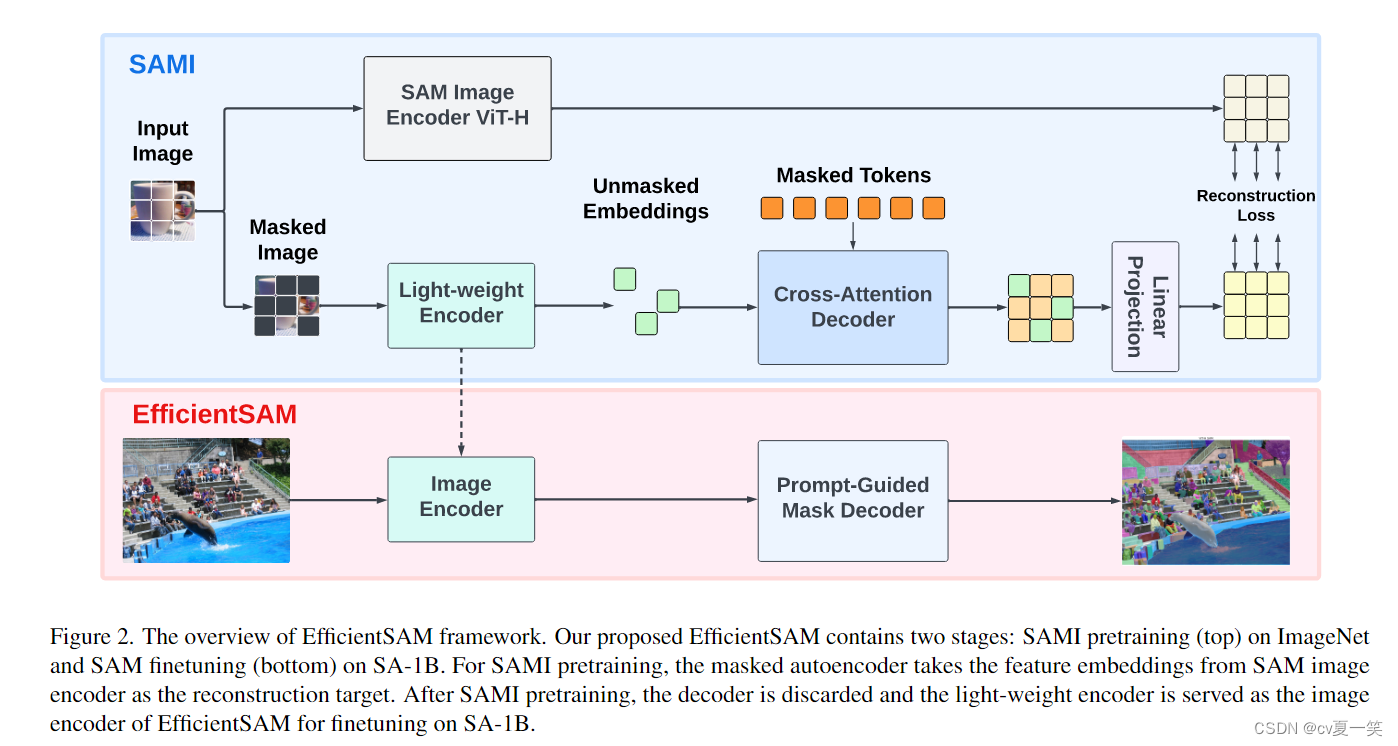
SAMI预训练(上半部分)在ImageNet上进行,而SAM微调(下半部分)则在SA-1B数据集上进行。
上半部分作用?
上半部分是为了得到轻量级编码器,因为sam本身的笨重的编码器是架构复杂推理慢的根本原因。
上半部分采用的方法
图像掩码预训练和损失重建。轻量级编码器学习重构来自于SAM VIT-H的图像编码器的特征嵌入,这样做可以使得很小的编码器也能输出和SAM VIT-H一样的编码特征。这个过程算是只是蒸馏,小模型学习目标是大模型的软输出,一个较大的“教师”模型向一个较小的“学生”模型传递知识。
下半部分作用?
上半部分训练了一个和VIT-H一样表现的模型,但是他并不具备segment anything的能力,meta的好多产品都是数据赋予模型能力,所以还是得加大数据集,之后在小模型上用大数据训练==赋能。同时训练了提示解码器,这里的提示可以点可以框。
EfficientSAM的结果
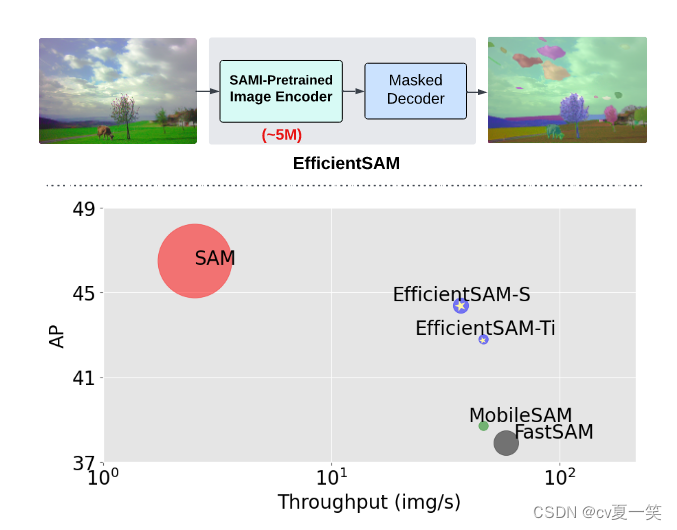
EfficientSAM-S 将 SAM 的推理时间减少了约 20 倍,参数大小减少了约 20 倍,性能略有下降,为 44.4 AP ,sam为 46.5 AP。
同时与近期工作对比中,约4AP高于MobileSAM FastSAM,而且处理复杂度差不多。
2️⃣EfficientSAM用于自己的数据集代码
微调的方式太多啦,比如只需要其编码器作为提特征的主干模块后面接自己的分类分割下游任务,这个我也写了但是暂时不考虑公开。比如需要全部的模型架构,但是自己数据集的域不同,所以要全量微调。比如将sam的主干和其他主干做模型融合/特征融合。方式很多,我们这里介绍最简单的,也是其他一切fashion变形的基础。
我们这里介绍全量微调,没加任何的fine-tune方法,只是全量更新模型参数适配自己的数据集。
提示选择的一个框提示。
训练方式有单卡和多卡。
处理数据集
每个人的数据存储方式和格式都不一样,前面几篇有处理成npy的代码可以参考,上一篇有根据路径编写Dataset.py的代码可以参考。因为我的数据量比较大,需要一个高效的存取方式,没过一万长的,随便存就行,自己写个Dataset.py 能输出img和label张量就行。
这里给到一个npy存储格式构造Dataset的代码,以供参考,实际根据自己的数据集情况写一个能对上模型输入就行。
特别注意一下img label bboxes boxes_labels的维度就可以
class NpyDataset(Dataset):def __init__(self, data_root, image_size=256, bbox_shift=5, data_aug=True):self.data_root = data_rootself.gt_path = join(data_root, 'gts')self.img_path = join(data_root, 'imgs')self.gt_path_files = sorted(glob(join(self.gt_path, '*.npy'), recursive=True))self.gt_path_files = [file for file in self.gt_path_filesif isfile(join(self.img_path, basename(file)))]self.image_size = image_sizeself.target_length = image_sizeself.bbox_shift = bbox_shiftself.data_aug = data_augdef __len__(self):return len(self.gt_path_files)def __getitem__(self, index):img_name = basename(self.gt_path_files[index])assert img_name == basename(self.gt_path_files[index]), 'img gt name error' + self.gt_path_files[index] + \self.npy_files[index]img_3c = np.load(join(self.img_path, img_name), 'r', allow_pickle=True) # (H, W, 3)img_resize = self.resize_longest_side(img_3c)# Resizingimg_resize = (img_resize - img_resize.min()) / np.clip(img_resize.max() - img_resize.min(), a_min=1e-8,a_max=None) # normalize to [0, 1], (H, W, 3img_padded = self.pad_image(img_resize) # (256, 256, 3)# convert the shape to (3, H, W)img_padded = np.transpose(img_padded, (2, 0, 1)) # (3, 256, 256)assert np.max(img_padded) <= 1.0 and np.min(img_padded) >= 0.0, 'image should be normalized to [0, 1]'gt = np.load(self.gt_path_files[index], 'r', allow_pickle=True) # multiple labels [0, 1,4,5...], (256,256)gt = cv2.resize(gt,(img_resize.shape[1], img_resize.shape[0]),interpolation=cv2.INTER_NEAREST).astype(np.uint8)gt = self.pad_image(gt) # (256, 256)label_ids = np.unique(gt)[1:]try:gt2D = np.uint8(gt == random.choice(label_ids.tolist())) # only one label, (256, 256)except:print(img_name, 'label_ids.tolist()', label_ids.tolist())gt2D = np.uint8(gt == np.max(gt)) # only one label, (256, 256)# add data augmentation: random fliplr and random flipudif self.data_aug:if random.random() > 0.5:img_padded = np.ascontiguousarray(np.flip(img_padded, axis=-1))gt2D = np.ascontiguousarray(np.flip(gt2D, axis=-1))# print('DA with flip left right')if random.random() > 0.5:img_padded = np.ascontiguousarray(np.flip(img_padded, axis=-2))gt2D = np.ascontiguousarray(np.flip(gt2D, axis=-2))# print('DA with flip upside down')gt2D = np.uint8(gt2D > 0)y_indices, x_indices = np.where(gt2D > 0)x_min, x_max = np.min(x_indices), np.max(x_indices)y_min, y_max = np.min(y_indices), np.max(y_indices)# add perturbation to bounding box coordinatesH, W = gt2D.shapex_min = max(0, x_min - random.randint(0, self.bbox_shift))x_max = min(W, x_max + random.randint(0, self.bbox_shift))y_min = max(0, y_min - random.randint(0, self.bbox_shift))y_max = min(H, y_max + random.randint(0, self.bbox_shift))# bboxes = np.array([x_min, y_min, x_max, y_max])bboxes = np.array([[x_min, y_min], [x_max, y_max]])boxes_1 = torch.reshape(torch.tensor(bboxes), [1, 1, -1, 2])input_label = np.array([2, 3])boxes_1 = torch.tensor(bboxes)[None, :] # boxes_1形状为[1, 2, 2]pts_labels = torch.tensor(input_label)[None, :] # pts_labels形状为[1, 2]return {"image": torch.tensor(img_padded).float(),"gt2D": torch.tensor(gt2D[None, :, :]).long(),# "bboxes": torch.tensor(bboxes[None, None, ...]).float(), # (B, 1, 4)"bboxes": boxes_1, # efficient模型需要这样的维度 (B, 1, 2, 2)"boxes_labels": pts_labels,"image_name": img_name,"new_size": torch.tensor(np.array([img_resize.shape[0], img_resize.shape[1]])).long(),"original_size": torch.tensor(np.array([img_3c.shape[0], img_3c.shape[1]])).long()}def resize_longest_side(self, image):"""Expects a numpy array with shape HxWxC in uint8 format."""long_side_length = self.target_lengtholdh, oldw = image.shape[0], image.shape[1]scale = long_side_length * 1.0 / max(oldh, oldw)newh, neww = oldh * scale, oldw * scaleneww, newh = int(neww + 0.5), int(newh + 0.5)target_size = (neww, newh)return cv2.resize(image, target_size, interpolation=cv2.INTER_AREA)def pad_image(self, image):"""Expects a numpy array with shape HxWxC in uint8 format."""# Padh, w = image.shape[0], image.shape[1]padh = self.image_size - hpadw = self.image_size - wif len(image.shape) == 3: ## Pad imageimage_padded = np.pad(image, ((0, padh), (0, padw), (0, 0)))else: ## Pad gt maskimage_padded = np.pad(image, ((0, padh), (0, padw)))return image_padded
将Efficient-SAM代码和权重拷贝到服务器或者本地
- 新建一个Net文件夹,将efficient-sam放下面
- 复制下面代码放Net下面
small_efficient_sam_encoder_config.py
# from Dataset.Dataset import train_loader
from Net.efficient_sam.efficient_sam_encoder import ImageEncoderViT
from torch import nn, Tensorimg_size = 1024
encoder_patch_size = 16
encoder_depth = 12
encoder_mlp_ratio = 4.0
encoder_neck_dims = [256, 256]
decoder_max_num_input_points = 6
decoder_transformer_depth = 2
decoder_transformer_mlp_dim = 2048
decoder_num_heads = 8
decoder_upscaling_layer_dims = [64, 32]
num_multimask_outputs = 3
iou_head_depth = 3
iou_head_hidden_dim = 256
activation = "gelu"
normalization_type = "layer_norm"
normalize_before_activation = False
small_efficient_sam_encoder = ImageEncoderViT(img_size=img_size,patch_size=encoder_patch_size,in_chans=3,# small vitpatch_embed_dim=384,normalization_type=normalization_type,depth=encoder_depth,# small vitnum_heads=6,mlp_ratio=encoder_mlp_ratio,neck_dims=encoder_neck_dims,act_layer=nn.ReLU,
)tiny_efficient_sam_encoder_config.py
# from Dataset.Dataset import train_loader
from Net.efficient_sam.efficient_sam_encoder import ImageEncoderViT
from torch import nn, Tensorimg_size = 1024
encoder_patch_size = 16
encoder_depth = 12
encoder_mlp_ratio = 4.0
encoder_neck_dims = [256, 256]
decoder_max_num_input_points = 6
decoder_transformer_depth = 2
decoder_transformer_mlp_dim = 2048
decoder_num_heads = 8
decoder_upscaling_layer_dims = [64, 32]
num_multimask_outputs = 3
iou_head_depth = 3
iou_head_hidden_dim = 256
activation = "gelu"
normalization_type = "layer_norm"
normalize_before_activation = False
tiny_efficient_sam_encoder = ImageEncoderViT(img_size=img_size,patch_size=encoder_patch_size,in_chans=3,# small vitpatch_embed_dim=192,normalization_type=normalization_type,depth=encoder_depth,# small vitnum_heads=3,mlp_ratio=encoder_mlp_ratio,neck_dims=encoder_neck_dims,act_layer=nn.ReLU,
)去官网git下载权重
放到项目下面,新建weights文件夹,只需要下载这两个就行,下载之后把zip解压
愉快训练
这里有一些参数需要根据自己的实际情况更改
单卡
# %%
import os
import random
import monai
from os import listdir, makedirs
from os.path import join, exists, isfile, isdir, basename
from glob import glob
from tqdm import tqdm, trange
from copy import deepcopy
from time import time
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader
from datetime import datetimeimport cv2
import torch.nn.functional as Ffrom matplotlib import pyplot as plt
import argparse# %%
parser = argparse.ArgumentParser()
parser.add_argument("-data_root", type=str, default="train_npy",help="Path to the npy data root."
)
parser.add_argument("-pretrained_checkpoint", type=str, default="lite_medsam.pth",help="Path to the pretrained Lite-MedSAM checkpoint."
)parser.add_argument("-work_dir", type=str, default="./workdir",help="Path to the working directory where checkpoints and logs will be saved."
)
parser.add_argument("-num_epochs", type=int, default=10,help="Number of epochs to train."
)
parser.add_argument("-batch_size", type=int, default=4,help="Batch size."
)
parser.add_argument("-num_workers", type=int, default=8,help="Number of workers for dataloader."
)
parser.add_argument("-device", type=str, default="cuda:1",help="Device to train on."
)
parser.add_argument("-bbox_shift", type=int, default=5,help="Perturbation to bounding box coordinates during training."
)
parser.add_argument("-lr", type=float, default=0.00005,help="Learning rate."
)
parser.add_argument("-weight_decay", type=float, default=0.01,help="Weight decay."
)
parser.add_argument("-iou_loss_weight", type=float, default=1.0,help="Weight of IoU loss."
)
parser.add_argument("-seg_loss_weight", type=float, default=1.0,help="Weight of segmentation loss."
)
parser.add_argument("-ce_loss_weight", type=float, default=1.0,help="Weight of cross entropy loss."
)
parser.add_argument("--sanity_check", action="store_true",help="Whether to do sanity check for dataloading."
)args = parser.parse_args()
# %%
work_dir = args.work_dir
data_root = args.data_root
medsam_lite_checkpoint = args.pretrained_checkpoint
num_epochs = args.num_epochs
batch_size = args.batch_size
num_workers = args.num_workers
device = args.device
bbox_shift = args.bbox_shift
lr = args.lr
weight_decay = args.weight_decay
iou_loss_weight = args.iou_loss_weight
seg_loss_weight = args.seg_loss_weight
ce_loss_weight = args.ce_loss_weight
do_sancheck = args.sanity_check
checkpoint = args.resumemakedirs(work_dir, exist_ok=True)# %%
torch.cuda.empty_cache()
os.environ["OMP_NUM_THREADS"] = "4" # export OMP_NUM_THREADS=4
os.environ["OPENBLAS_NUM_THREADS"] = "4" # export OPENBLAS_NUM_THREADS=4
os.environ["MKL_NUM_THREADS"] = "6" # export MKL_NUM_THREADS=6
os.environ["VECLIB_MAXIMUM_THREADS"] = "4" # export VECLIB_MAXIMUM_THREADS=4
os.environ["NUMEXPR_NUM_THREADS"] = "6" # export NUMEXPR_NUM_THREADS=6def show_mask(mask, ax, random_color=False):if random_color:color = np.concatenate([np.random.random(3), np.array([0.45])], axis=0)else:color = np.array([251 / 255, 252 / 255, 30 / 255, 0.45])h, w = mask.shape[-2:]mask_image = mask.reshape(h, w, 1) * color.reshape(1, 1, -1)ax.imshow(mask_image)def show_box(box, ax):x0, y0 = box[0], box[1]w, h = box[2] - box[0], box[3] - box[1]ax.add_patch(plt.Rectangle((x0, y0), w, h, edgecolor='blue', facecolor=(0, 0, 0, 0), lw=2))def cal_iou(result, reference):intersection = torch.count_nonzero(torch.logical_and(result, reference), dim=[i for i in range(1, result.ndim)])union = torch.count_nonzero(torch.logical_or(result, reference), dim=[i for i in range(1, result.ndim)])iou = intersection.float() / union.float()return iou.unsqueeze(1)# %%
class NpyDataset(Dataset):def __init__(self, data_root, image_size=256, bbox_shift=5, data_aug=True):self.data_root = data_rootself.gt_path = join(data_root, 'gts')self.img_path = join(data_root, 'imgs')self.gt_path_files = sorted(glob(join(self.gt_path, '*.npy'), recursive=True))self.gt_path_files = [file for file in self.gt_path_filesif isfile(join(self.img_path, basename(file)))]self.image_size = image_sizeself.target_length = image_sizeself.bbox_shift = bbox_shiftself.data_aug = data_augdef __len__(self):return len(self.gt_path_files)def __getitem__(self, index):img_name = basename(self.gt_path_files[index])assert img_name == basename(self.gt_path_files[index]), 'img gt name error' + self.gt_path_files[index] + \self.npy_files[index]img_3c = np.load(join(self.img_path, img_name), 'r', allow_pickle=True) # (H, W, 3)img_resize = self.resize_longest_side(img_3c)# Resizingimg_resize = (img_resize - img_resize.min()) / np.clip(img_resize.max() - img_resize.min(), a_min=1e-8,a_max=None) # normalize to [0, 1], (H, W, 3img_padded = self.pad_image(img_resize) # (256, 256, 3)# convert the shape to (3, H, W)img_padded = np.transpose(img_padded, (2, 0, 1)) # (3, 256, 256)assert np.max(img_padded) <= 1.0 and np.min(img_padded) >= 0.0, 'image should be normalized to [0, 1]'gt = np.load(self.gt_path_files[index], 'r', allow_pickle=True) # multiple labels [0, 1,4,5...], (256,256)gt = cv2.resize(gt,(img_resize.shape[1], img_resize.shape[0]),interpolation=cv2.INTER_NEAREST).astype(np.uint8)gt = self.pad_image(gt) # (256, 256)label_ids = np.unique(gt)[1:]try:gt2D = np.uint8(gt == random.choice(label_ids.tolist())) # only one label, (256, 256)except:print(img_name, 'label_ids.tolist()', label_ids.tolist())gt2D = np.uint8(gt == np.max(gt)) # only one label, (256, 256)# add data augmentation: random fliplr and random flipudif self.data_aug:if random.random() > 0.5:img_padded = np.ascontiguousarray(np.flip(img_padded, axis=-1))gt2D = np.ascontiguousarray(np.flip(gt2D, axis=-1))# print('DA with flip left right')if random.random() > 0.5:img_padded = np.ascontiguousarray(np.flip(img_padded, axis=-2))gt2D = np.ascontiguousarray(np.flip(gt2D, axis=-2))# print('DA with flip upside down')gt2D = np.uint8(gt2D > 0)y_indices, x_indices = np.where(gt2D > 0)x_min, x_max = np.min(x_indices), np.max(x_indices)y_min, y_max = np.min(y_indices), np.max(y_indices)# add perturbation to bounding box coordinatesH, W = gt2D.shapex_min = max(0, x_min - random.randint(0, self.bbox_shift))x_max = min(W, x_max + random.randint(0, self.bbox_shift))y_min = max(0, y_min - random.randint(0, self.bbox_shift))y_max = min(H, y_max + random.randint(0, self.bbox_shift))# bboxes = np.array([x_min, y_min, x_max, y_max])bboxes = np.array([[x_min, y_min], [x_max, y_max]])boxes_1 = torch.reshape(torch.tensor(bboxes), [1, 1, -1, 2])input_label = np.array([2, 3])boxes_1 = torch.tensor(bboxes)[None, :] # boxes_1形状为[1, 2, 2]pts_labels = torch.tensor(input_label)[None, :] # pts_labels形状为[1, 2]return {"image": torch.tensor(img_padded).float(),"gt2D": torch.tensor(gt2D[None, :, :]).long(),# "bboxes": torch.tensor(bboxes[None, None, ...]).float(), # (B, 1, 4)"bboxes": boxes_1, # efficient模型需要这样的维度 (B, 1, 2, 2)"boxes_labels": pts_labels,"image_name": img_name,"new_size": torch.tensor(np.array([img_resize.shape[0], img_resize.shape[1]])).long(),"original_size": torch.tensor(np.array([img_3c.shape[0], img_3c.shape[1]])).long()}def resize_longest_side(self, image):"""Expects a numpy array with shape HxWxC in uint8 format."""long_side_length = self.target_lengtholdh, oldw = image.shape[0], image.shape[1]scale = long_side_length * 1.0 / max(oldh, oldw)newh, neww = oldh * scale, oldw * scaleneww, newh = int(neww + 0.5), int(newh + 0.5)target_size = (neww, newh)return cv2.resize(image, target_size, interpolation=cv2.INTER_AREA)def pad_image(self, image):"""Expects a numpy array with shape HxWxC in uint8 format."""# Padh, w = image.shape[0], image.shape[1]padh = self.image_size - hpadw = self.image_size - wif len(image.shape) == 3: ## Pad imageimage_padded = np.pad(image, ((0, padh), (0, padw), (0, 0)))else: ## Pad gt maskimage_padded = np.pad(image, ((0, padh), (0, padw)))return image_padded# %% sanity test of dataset class
if do_sancheck:tr_dataset = NpyDataset(data_root, data_aug=True)tr_dataloader = DataLoader(tr_dataset, batch_size=8, shuffle=True)for step, batch in enumerate(tr_dataloader):# show the example_, axs = plt.subplots(1, 2, figsize=(10, 10))idx = random.randint(0, 4)image = batch["image"]gt = batch["gt2D"]bboxes = batch["bboxes"]names_temp = batch["image_name"]axs[0].imshow(image[idx].cpu().permute(1, 2, 0).numpy())show_mask(gt[idx].cpu().squeeze().numpy(), axs[0])show_box(bboxes[idx].numpy().squeeze(), axs[0])axs[0].axis('off')# set titleaxs[0].set_title(names_temp[idx])idx = random.randint(4, 7)axs[1].imshow(image[idx].cpu().permute(1, 2, 0).numpy())show_mask(gt[idx].cpu().squeeze().numpy(), axs[1])show_box(bboxes[idx].numpy().squeeze(), axs[1])axs[1].axis('off')# set titleaxs[1].set_title(names_temp[idx])plt.subplots_adjust(wspace=0.01, hspace=0)plt.savefig(join(work_dir, 'medsam_lite-train_bbox_prompt_sanitycheck_DA.png'),bbox_inches='tight',dpi=300)plt.close()break# %%
class MedSAM_Lite(nn.Module):def __init__(self,image_encoder,mask_decoder,prompt_encoder):super().__init__()self.image_encoder = image_encoderself.mask_decoder = mask_decoderself.prompt_encoder = prompt_encoderdef forward(self, image, boxes):image_embedding = self.image_encoder(image) # (B, 256, 64, 64)sparse_embeddings, dense_embeddings = self.prompt_encoder(points=None,boxes=boxes,masks=None,)low_res_masks, iou_predictions = self.mask_decoder(image_embeddings=image_embedding, # (B, 256, 64, 64)image_pe=self.prompt_encoder.get_dense_pe(), # (1, 256, 64, 64)sparse_prompt_embeddings=sparse_embeddings, # (B, 2, 256)dense_prompt_embeddings=dense_embeddings, # (B, 256, 64, 64)multimask_output=False,) # (B, 1, 256, 256)return low_res_masks, iou_predictions@torch.no_grad()def postprocess_masks(self, masks, new_size, original_size):"""Do cropping and resizing"""# Cropmasks = masks[:, :, :new_size[0], :new_size[1]]# Resizemasks = F.interpolate(masks,size=(original_size[0], original_size[1]),mode="bilinear",align_corners=False,)return masksfrom Net.efficient_sam.build_efficient_sam import build_efficient_sam_vits
medsam_lite_model = build_efficient_sam_vits()
medsam_lite_model = medsam_lite_model.to(device)
medsam_lite_model.train()# %%
print(f"MedSAM Lite size: {sum(p.numel() for p in medsam_lite_model.parameters())}")
# %%
optimizer = optim.AdamW(medsam_lite_model.parameters(),lr=lr,betas=(0.9, 0.999),eps=1e-08,weight_decay=weight_decay,
)
lr_scheduler = optim.lr_scheduler.ReduceLROnPlateau(optimizer,mode='min',factor=0.9,patience=5,cooldown=0
)
seg_loss = monai.losses.DiceLoss(sigmoid=True, squared_pred=True, reduction='mean')
ce_loss = nn.BCEWithLogitsLoss(reduction='mean')
iou_loss = nn.MSELoss(reduction='mean')
# %%
train_dataset = NpyDataset(data_root=data_root, data_aug=True)
train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True, num_workers=num_workers, pin_memory=True)if checkpoint and isfile(checkpoint):print(f"Resuming from checkpoint {checkpoint}")checkpoint = torch.load(checkpoint)medsam_lite_model.load_state_dict(checkpoint["model"], strict=True)optimizer.load_state_dict(checkpoint["optimizer"])start_epoch = checkpoint["epoch"]best_loss = checkpoint["loss"]print(f"Loaded checkpoint from epoch {start_epoch}")
else:start_epoch = 0best_loss = 1e10
# %%
train_losses = []
for epoch in range(start_epoch + 1, num_epochs):epoch_loss = [1e10 for _ in range(len(train_loader))]epoch_start_time = time()pbar = tqdm(train_loader)for step, batch in enumerate(pbar):image = batch["image"]gt2D = batch["gt2D"]boxes = batch["bboxes"]label_box = batch["boxes_labels"]optimizer.zero_grad()image, gt2D, boxes, label_box = image.to(device), gt2D.to(device), boxes.to(device), label_box.to(device)logits_pred, iou_pred = medsam_lite_model(image, boxes, label_box)gt2D = torch.unsqueeze(gt2D, 2)gt2D = gt2D.repeat(1, 1, 3, 1, 1)l_seg = seg_loss(logits_pred, gt2D)l_ce = ce_loss(logits_pred, gt2D.float())# mask_loss = l_seg + l_cemask_loss = seg_loss_weight * l_seg + ce_loss_weight * l_ceiou_gt = cal_iou(torch.sigmoid(logits_pred) > 0.5, gt2D.bool())l_iou = iou_loss(iou_pred, iou_gt)# loss = mask_loss + l_iouloss = mask_loss + iou_loss_weight * l_iouepoch_loss[step] = loss.item()loss.backward()optimizer.step()optimizer.zero_grad()pbar.set_description(f"Epoch {epoch} at {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}, loss: {loss.item():.4f}")epoch_end_time = time()epoch_loss_reduced = sum(epoch_loss) / len(epoch_loss)train_losses.append(epoch_loss_reduced)lr_scheduler.step(epoch_loss_reduced)model_weights = medsam_lite_model.state_dict()checkpoint = {"model": model_weights,"epoch": epoch,"optimizer": optimizer.state_dict(),"loss": epoch_loss_reduced,"best_loss": best_loss,}torch.save(checkpoint, join(work_dir, "medsam_lite_latest.pth"))if epoch_loss_reduced < best_loss:print(f"New best loss: {best_loss:.4f} -> {epoch_loss_reduced:.4f}")best_loss = epoch_loss_reducedcheckpoint["best_loss"] = best_losstorch.save(checkpoint, join(work_dir, "medsam_lite_best.pth"))epoch_loss_reduced = 1e10# %% plot lossplt.plot(train_losses)plt.title("Dice + Binary Cross Entropy + IoU Loss")plt.xlabel("Epoch")plt.ylabel("Loss")plt.savefig(join(work_dir, "train_loss.png"))plt.close()多卡
我这里有其他团队成员在用卡,所以此时此刻一个rank我只能用2,3号gpu。根据自己情况更改
# %%
import os
import random
import monai
from os import listdir, makedirs
from os.path import join, isfile, basename
from glob import glob
from tqdm import tqdm
from copy import deepcopy
from time import time
from shutil import copyfile
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader
from torch import multiprocessing as mp
from torch import distributed as dist
from datetime import datetime
import cv2
import torch.nn.functional as Ffrom matplotlib import pyplot as plt
import argparse
import torch# print(torch.cuda.nccl.version())torch.cuda.empty_cache()
os.environ["OMP_NUM_THREADS"] = "4" # export OMP_NUM_THREADS=4
os.environ["OPENBLAS_NUM_THREADS"] = "4" # export OPENBLAS_NUM_THREADS=4
os.environ["MKL_NUM_THREADS"] = "6" # export MKL_NUM_THREADS=6
os.environ["VECLIB_MAXIMUM_THREADS"] = "4" # export VECLIB_MAXIMUM_THREADS=4
os.environ["NUMEXPR_NUM_THREADS"] = "1" # export NUMEXPR_NUM_THREADS=6
os.environ['MASTER_ADDR'] = '' # IP of node with rank 0
os.environ['MASTER_PORT'] = '' # Port on master node
os.environ['WORLD_SIZE'] = '2' # Total number of processes
os.environ['RANK'] = '0' # Rank of this processdef get_args():parser = argparse.ArgumentParser()parser.add_argument('-i', '--tr_npy_path', type=str,default='train_npy',help='Path to training npy files; two subfolders: gts and imgs')parser.add_argument('-task_name', type=str, default='MedSAM-Lite')parser.add_argument('-pretrained_checkpoint', type=str,help='Path to pretrained MedSAM-Lite checkpoint')parser.add_argument('-work_dir', type=str, default='./work_dir_multi')parser.add_argument('--data_aug', action='store_true', default=False,help='use data augmentation during training')# trainparser.add_argument('-num_epochs', type=int, default=1000)parser.add_argument('-batch_size', type=int, default=4)parser.add_argument('-num_workers', type=int, default=1)# Optimizer parametersparser.add_argument('-weight_decay', type=float, default=0.01,help='weight decay (default: 0.01)')parser.add_argument('-lr', type=float, default=0.0005, metavar='LR',help='learning rate (absolute lr)')## Distributed training argsparser.add_argument('-world_size', type=int, default=2, help='world size')parser.add_argument('-node_rank', default=0, type=int, help='Node rank')parser.add_argument('-bucket_cap_mb', type=int, default=25,help='The amount of memory in Mb that DDP will accumulate before firing off gradient communication for the bucket (need to tune)')parser.add_argument('-resume', type=str, default='', required=False,help="Resuming training from a work_dir")parser.add_argument('-init_method', type=str, defauargs = parser.parse_args()return argsdef show_mask(mask, ax, random_color=False):if random_color:color = np.concatenate([np.random.random(3), np.array([0.45])], axis=0)else:color = np.array([251 / 255, 252 / 255, 30 / 255, 0.45])h, w = mask.shape[-2:]mask_image = mask.reshape(h, w, 1) * color.reshape(1, 1, -1)ax.imshow(mask_image)def show_box(box, ax):x0, y0 = box[0], box[1]w, h = box[2] - box[0], box[3] - box[1]ax.add_patch(plt.Rectangle((x0, y0), w, h, edgecolor='blue', facecolor=(0, 0, 0, 0), lw=2))@torch.no_grad()
def cal_iou(result, reference):intersection = torch.count_nonzero(torch.logical_and(result, reference), dim=[i for i in range(1, result.ndim)])union = torch.count_nonzero(torch.logical_or(result, reference), dim=[i for i in range(1, result.ndim)])iou = intersection.float() / union.float()return iou.unsqueeze(1)def revert_sync_batchnorm(module: torch.nn.Module) -> torch.nn.Module:# Code adapted from https://github.com/pytorch/pytorch/issues/41081#issuecomment-783961547# Original author: Kapil Yedidi (@kapily)converted_module = moduleif isinstance(module, torch.nn.modules.batchnorm.SyncBatchNorm):# Unfortunately, SyncBatchNorm does not store the original class - if it did# we could return the one that was originally created.converted_module = nn.BatchNorm2d(module.num_features, module.eps, module.momentum, module.affine, module.track_running_stats)if module.affine:with torch.no_grad():converted_module.weight = module.weightconverted_module.bias = module.biasconverted_module.running_mean = module.running_meanconverted_module.running_var = module.running_varconverted_module.num_batches_tracked = module.num_batches_trackedif hasattr(module, "qconfig"):converted_module.qconfig = module.qconfigfor name, child in module.named_children():converted_module.add_module(name, revert_sync_batchnorm(child))del modulereturn converted_moduleclass NpyDataset(Dataset):def __init__(self, data_root, image_size=256, bbox_shift=10, data_aug=True):self.data_root = data_rootself.gt_path = join(data_root, 'gts')self.img_path = join(data_root, 'imgs')self.gt_path_files = sorted(glob(join(self.gt_path, '*.npy'), recursive=True))self.gt_path_files = [file for file in self.gt_path_files if isfile(join(self.img_path, basename(file)))]self.image_size = image_sizeself.target_length = image_sizeself.bbox_shift = bbox_shiftself.data_aug = data_augdef __len__(self):return len(self.gt_path_files)def __getitem__(self, index):img_name = basename(self.gt_path_files[index])assert img_name == basename(self.gt_path_files[index]), 'img gt name error' + self.gt_path_files[index] + \self.npy_files[index]img_3c = np.load(join(self.img_path, img_name), 'r', allow_pickle=True) # (H, W, 3)# Resizing and normalizationimg_resize = self.resize_longest_side(img_3c)img_resize = (img_resize - img_resize.min()) / np.clip(img_resize.max() - img_resize.min(), a_min=1e-8,a_max=None) # normalize to [0, 1], (H, W, 3img_padded = self.pad_image(img_resize) # (256, 256, 3)# convert the shape to (3, H, W)img_padded = np.transpose(img_padded, (2, 0, 1)) # (3, 256, 256)assert np.max(img_padded) <= 1.0 and np.min(img_padded) >= 0.0, 'image should be normalized to [0, 1]'gt = np.load(self.gt_path_files[index], 'r', allow_pickle=True) # multiple labels [0, 1,4,5...], (256,256)assert gt.max() >= 1, 'gt should have at least one label'gt = cv2.resize(gt,(img_resize.shape[1], img_resize.shape[0]),interpolation=cv2.INTER_NEAREST).astype(np.uint8)gt = self.pad_image(gt) # (256, 256)label_ids = np.unique(gt)[1:]try:gt2D = np.uint8(gt == random.choice(label_ids.tolist())) # only one label, (256, 256)except:print(img_name, 'label_ids.tolist()', label_ids.tolist())gt2D = np.uint8(gt == np.max(gt)) # only one label, (256, 256)# add data augmentation: random fliplr and random flipudif self.data_aug:if random.random() > 0.5:img_padded = np.ascontiguousarray(np.flip(img_padded, axis=-1))gt2D = np.ascontiguousarray(np.flip(gt2D, axis=-1))# print('DA with flip left right')if random.random() > 0.5:img_padded = np.ascontiguousarray(np.flip(img_padded, axis=-2))gt2D = np.ascontiguousarray(np.flip(gt2D, axis=-2))# print('DA with flip upside down')gt2D = np.uint8(gt2D > 0)y_indices, x_indices = np.where(gt2D > 0)x_min, x_max = np.min(x_indices), np.max(x_indices)y_min, y_max = np.min(y_indices), np.max(y_indices)# add perturbation to bounding box coordinatesH, W = gt2D.shapex_min = max(0, x_min - random.randint(0, self.bbox_shift))x_max = min(W, x_max + random.randint(0, self.bbox_shift))y_min = max(0, y_min - random.randint(0, self.bbox_shift))y_max = min(H, y_max + random.randint(0, self.bbox_shift))# bboxes = np.array([x_min, y_min, x_max, y_max])bboxes = np.array([[x_min, y_min], [x_max, y_max]])input_label = np.array([2, 3])# pts_labels = torch.reshape(torch.tensor(input_label), [1, 1, -1])# bboxes已经是[2, 2]形状,我们只需要增加一个批次维度boxes_1 = torch.tensor(bboxes)[None, :] # boxes_1形状为[1, 2, 2]# input_label是[2]形状,我们也是增加一个批次维度pts_labels = torch.tensor(input_label)[None, :] # pts_labels形状为[1, 2]return {"image": torch.tensor(img_padded).float(),"gt2D": torch.tensor(gt2D[None, :, :]).long(),# "bboxes": torch.tensor(bboxes[None, None, ...]).float(), # (B, 1, 4)"bboxes": boxes_1, # efficient模型需要这样的维度 (B, 1, 2, 2)"boxes_labels": pts_labels,"image_name": img_name,"new_size": torch.tensor(np.array([img_resize.shape[0], img_resize.shape[1]])).long(),"original_size": torch.tensor(np.array([img_3c.shape[0], img_3c.shape[1]])).long()}def resize_longest_side(self, image):"""Expects a numpy array with shape HxWxC in uint8 format."""long_side_length = self.target_lengtholdh, oldw = image.shape[0], image.shape[1]scale = long_side_length * 1.0 / max(oldh, oldw)newh, neww = oldh * scale, oldw * scaleneww, newh = int(neww + 0.5), int(newh + 0.5)target_size = (neww, newh)return cv2.resize(image, target_size, interpolation=cv2.INTER_AREA)def pad_image(self, image):"""Expects a numpy array with shape HxWxC in uint8 format."""# Padh, w = image.shape[0], image.shape[1]padh = self.image_size - hpadw = self.image_size - wif len(image.shape) == 3: ## Pad imageimage_padded = np.pad(image, ((0, padh), (0, padw), (0, 0)))else: ## Pad gt maskimage_padded = np.pad(image, ((0, padh), (0, padw)))return image_paddeddef collate_fn(batch):"""Collate function for PyTorch DataLoader."""batch_dict = {}for key in batch[0].keys():if key == "image_name":batch_dict[key] = [sample[key] for sample in batch]else:batch_dict[key] = torch.stack([sample[key] for sample in batch], dim=0)return batch_dict# %% sanity test of dataset class
def sanity_check_dataset(args):print('tr_npy_path', args.tr_npy_path)tr_dataset = NpyDataset(args.tr_npy_path, data_aug=args.data_aug)print('len(tr_dataset)', len(tr_dataset))tr_dataloader = DataLoader(tr_dataset, batch_size=4, shuffle=True, collate_fn=collate_fn)makedirs(args.work_dir, exist_ok=True)for step, batch in enumerate(tr_dataloader):# print(image.shape, gt.shape, bboxes.shape)# show the example_, axs = plt.subplots(1, 2, figsize=(10, 10))idx = random.randint(0, 4)image = batch["image"]gt = batch["gt2D"]bboxes = batch["bboxes"]names_temp = batch["image_name"]axs[0].imshow(image[idx].cpu().permute(1, 2, 0).numpy())show_mask(gt[idx].cpu().squeeze().numpy(), axs[0])show_box(bboxes[idx].numpy().squeeze(), axs[0])axs[0].axis('off')# set titleaxs[0].set_title(names_temp[idx])idx = random.randint(4, 7)axs[1].imshow(image[idx].cpu().permute(1, 2, 0).numpy())show_mask(gt[idx].cpu().squeeze().numpy(), axs[1])show_box(bboxes[idx].numpy().squeeze(), axs[1])axs[1].axis('off')# set titleaxs[1].set_title(names_temp[idx])# plt.show() plt.subplots_adjust(wspace=0.01, hspace=0)plt.savefig(join(args.work_dir, 'medsam_lite-train_bbox_prompt_sanitycheck_DA.png'),bbox_inches='tight',dpi=300)plt.close()break# %%
class MedSAM_Lite(nn.Module):def __init__(self,image_encoder,mask_decoder,prompt_encoder):super().__init__()self.image_encoder = image_encoderself.mask_decoder = mask_decoderself.prompt_encoder = prompt_encoderdef forward(self, image, boxes):image_embedding = self.image_encoder(image) # (B, 256, 64, 64)sparse_embeddings, dense_embeddings = self.prompt_encoder(points=None,boxes=boxes,masks=None,)low_res_logits, iou_predictions = self.mask_decoder(image_embeddings=image_embedding, # (B, 256, 64, 64)image_pe=self.prompt_encoder.get_dense_pe(), # (1, 256, 64, 64)sparse_prompt_embeddings=sparse_embeddings, # (B, 2, 256)dense_prompt_embeddings=dense_embeddings, # (B, 256, 64, 64)multimask_output=False,) # (B, 1, 256, 256)return low_res_logits, iou_predictions@torch.no_grad()def postprocess_masks(self, masks, new_size, original_size):"""Do cropping and resizing"""# Cropmasks = masks[:, :, :new_size[0], :new_size[1]]# Resizemasks = F.interpolate(masks,size=(original_size[0], original_size[1]),mode="bilinear",align_corners=False,)return masksdef main(args):ngpus_per_node = 2print("Spwaning processces")mp.spawn(main_worker, nprocs=ngpus_per_node, args=(ngpus_per_node, args))def main_worker(gpu, ngpus_per_node, args):node_rank = int(args.node_rank)adjusted_gpu = gpu + 1rank = node_rank * ngpus_per_node + adjusted_gpu-1# rank = node_rank * ngpus_per_node + gpuworld_size = args.world_size# print(f"[Rank {rank}]: Use GPU: {gpu} for training")print(f"[Rank {rank}]: Use GPU: {adjusted_gpu} for training")is_main_host = rank == 0print("now 1")if is_main_host:print("now 2")run_id = datetime.now().strftime("%Y%m%d-%H%M")model_save_path = join(args.work_dir, args.task_name + "-" + run_id)makedirs(model_save_path, exist_ok=True)copyfile(__file__, join(model_save_path, run_id + "_" + os.path.basename(__file__)))print("now 3")torch.cuda.set_device(adjusted_gpu)device = torch.device("cuda:{}".format(adjusted_gpu))print(device)print("now 4")dist.init_process_group(backend="nccl", init_method=args.init_method, rank=rank, world_size=world_size)print("now 5")num_epochs = args.num_epochsbatch_size = args.batch_sizenum_workers = args.num_workersfrom Net.efficient_sam.build_efficient_sam import build_efficient_sam_vitsprint("now 6")medsam_lite_model = build_efficient_sam_vits()medsam_lite_model = medsam_lite_model.to(device)## Make sure there's only 2d BN layers, so that I can revert them properlyfor module in medsam_lite_model.modules():cls_name = module.__class__.__name__if "BatchNorm" in cls_name:assert cls_name == "BatchNorm2d"medsam_lite_model = nn.SyncBatchNorm.convert_sync_batchnorm(medsam_lite_model)medsam_lite_model = nn.parallel.DistributedDataParallel(medsam_lite_model,device_ids=[adjusted_gpu],output_device=adjusted_gpu,find_unused_parameters=True,bucket_cap_mb=args.bucket_cap_mb)medsam_lite_model.train()# %%print(f"MedSAM Lite size: {sum(p.numel() for p in medsam_lite_model.parameters())}")# %%optimizer = optim.AdamW(medsam_lite_model.parameters(),lr=args.lr,betas=(0.9, 0.999),eps=1e-08,weight_decay=args.weight_decay,)lr_scheduler = optim.lr_scheduler.ReduceLROnPlateau(optimizer,mode='min',factor=0.9,patience=5,cooldown=0)seg_loss = monai.losses.DiceLoss(sigmoid=True, squared_pred=True, reduction='mean')ce_loss = nn.BCEWithLogitsLoss(reduction='mean')iou_loss = nn.MSELoss(reduction='mean')# %%data_root = args.tr_npy_pathtrain_dataset = NpyDataset(data_root=data_root, data_aug=True)train_sampler = torch.utils.data.distributed.DistributedSampler(train_dataset)train_loader = DataLoader(train_dataset,batch_size=batch_size,shuffle=False,num_workers=num_workers,pin_memory=True,sampler=train_sampler,collate_fn=collate_fn)# %%if os.path.exists(args.resume):ckpt_folders = sorted(listdir(args.resume))ckpt_folders = [f for f in ckpt_folders if(f.startswith(args.task_name) and isfile(join(args.resume, f, 'medsam_lite_latest.pth')))]print('*' * 20)print('existing ckpts in', args.resume, ckpt_folders)# find the latest ckpt folderstime_strings = [f.split(args.task_name + '-')[-1] for f in ckpt_folders]dates = [datetime.strptime(f, '%Y%m%d-%H%M') for f in time_strings]latest_date = max(dates)latest_ckpt = join(args.work_dir, args.task_name + '-' + latest_date.strftime('%Y%m%d-%H%M'),'medsam_lite_latest.pth')print('Loading from', latest_ckpt)checkpoint = torch.load(latest_ckpt, map_location=device)medsam_lite_model.module.load_state_dict(checkpoint["model"])optimizer.load_state_dict(checkpoint["optimizer"])start_epoch = checkpoint["epoch"] + 1best_loss = checkpoint["loss"]print(f"Loaded checkpoint from epoch {start_epoch}")else:start_epoch = 0best_loss = 1e10train_losses = []epoch_times = []for epoch in range(start_epoch, num_epochs):epoch_loss = [1e10 for _ in range(len(train_loader))]epoch_start_time = time()pbar = tqdm(train_loader)for step, batch in enumerate(pbar):image = batch["image"]gt2D = batch["gt2D"]boxes = batch["bboxes"]label_box = batch["boxes_labels"]optimizer.zero_grad()image, gt2D, boxes, label_box = image.to(device), gt2D.to(device), boxes.to(device), label_box.to(device)logits_pred, iou_pred = medsam_lite_model(image, boxes, label_box)gt2D = torch.unsqueeze(gt2D, 2)gt2D = gt2D.repeat(1, 1, 3, 1, 1)l_seg = seg_loss(logits_pred, gt2D)l_ce = ce_loss(logits_pred, gt2D.float())mask_loss = l_seg + l_cewith torch.no_grad():iou_gt = cal_iou(torch.sigmoid(logits_pred) > 0.5, gt2D.bool())l_iou = iou_loss(iou_pred, iou_gt)loss = mask_loss + l_iouepoch_loss[step] = loss.item()loss.backward()optimizer.step()optimizer.zero_grad()pbar.set_description(f"[RANK {rank}] Epoch {epoch} at {datetime.now().strftime('%Y-%m-%d %H:%M:%S')}, loss: {loss.item():.4f}")epoch_end_time = time()epoch_duration = epoch_end_time - epoch_start_timeepoch_times.append(epoch_duration)epoch_loss_world = [None for _ in range(world_size)]dist.all_gather_object(epoch_loss_world, epoch_loss)epoch_loss_reduced = np.vstack(epoch_loss_world).mean()train_losses.append(epoch_loss_reduced)lr_scheduler.step(epoch_loss_reduced)if is_main_host:module_revert_sync_BN = revert_sync_batchnorm(deepcopy(medsam_lite_model.module))weights = module_revert_sync_BN.state_dict()checkpoint = {"model": weights,"epoch": epoch,"optimizer": optimizer.state_dict(),"loss": epoch_loss_reduced,"best_loss": best_loss,}torch.save(checkpoint, join(model_save_path, "medsam_lite_latest.pth"))if epoch_loss_reduced < best_loss:print(f"New best loss: {best_loss:.4f} -> {epoch_loss_reduced:.4f}")best_loss = epoch_loss_reducedif is_main_host:checkpoint["best_loss"] = best_losstorch.save(checkpoint, join(model_save_path, "medsam_lite_best.pth"))dist.barrier()epoch_loss_reduced = 1e10# %% plot lossif is_main_host:fig, axes = plt.subplots(2, 1, figsize=(10, 8))axes[0].title.set_text("Dice + Binary Cross Entropy + IoU Loss")axes[0].plot(train_losses)axes[0].set_ylabel("Loss")axes[1].plot(epoch_times)axes[1].title.set_text("Epoch Duration")axes[1].set_ylabel("Duration (s)")axes[1].set_xlabel("Epoch")plt.tight_layout()plt.savefig(join(model_save_path, "log.png"))plt.close()dist.barrier()# %%
if __name__ == "__main__":args = get_args()# sanity_check_dataset(args)main(args)这篇关于【Segment Anything Model】十三:Meta的最新工作EfficientSAM,微调到自己的数据集,代码。的文章就介绍到这儿,希望我们推荐的文章对编程师们有所帮助!